
If you were adopted, would you want to know for certain who your biological parents are? If your family had a history of a particular disease, would you want to know your chances of getting it? These and other looming questions have major bioethical implications, but for years the answers were locked away in our DNA; always present, but impenetrable to decoding. As instruments and techniques were developed and optimized, researchers were finally able to sequence individual genes and study their biological roles. This advancing wave of technology combined with the completion of the human genome sequence in 2001 empowered scientists and physicians with the framework to make swift progress in the field of medicine via the analysis of our entire DNA code. Despite immense progress scientists have made, the use of advanced sequencing technology has been primarily confined to laboratories and unavailable to the general public. However, in the past few years several private biotech companies have started offering a "personal genome service" that involves sequencing the most variable portions of our DNA. The goals are straightforward - to give individuals information about their ancestry and inherited traits. While there are definite limitations - both technically and bioethically - to the amount and type of information that can be obtained from personal genome sequencing, in my case the service answered a lingering question about something important to me, and thus was well worth it.
In this article, I'm going to tell the story about why I chose to purchase a personal genome service, briefly explain how it works, show my interesting results, and finally, provide some commentary on how these services will impact the fields of genomics and medicine.
I have a twin brother named Matt. He and I look incredibly alike. I always believed that we were identical, and the majority of people who meet us agree. Therefore, you are likely to be puzzled upon reading that whenever anyone asked whether my Matt and I were identical, I would always respond "it's a long story". You see, at birth my mother's obstetrician told her that Matt and I were fraternal because my mother had two placentas. Without getting into the scientific details, it was conventional medical wisdom in 1986 that two placentas with twins meant they were fraternal. This was later determined to not be 100% true, which gave me hope that I indeed was an identical twin. But throughout my childhood, adolescence, and now adulthood, there always some was lingering doubt. As Matt and I grew older, we did begin to look a little bit different, especially in face shape. Now as adults, we started to get more comments from people saying they thought we were fraternal twins.
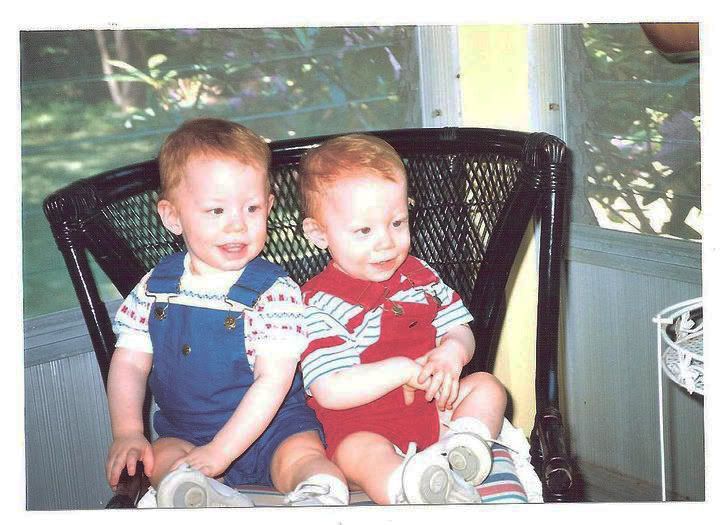
Baby Mike (left) and Matt (right). In addition to being extremely cute, we're indistinguishable!

Matt (right) and I (left) at age 9. Still looking very alike.

Matt (left) and I (right) this past year (age 24). Clearly still twins, but even I'll acknowledge some differences in face shape.
Now some of you are surely thinking "who cares if you are fraternal or identical!" My only response is that I think the true essence of being a twin is to be identical; otherwise, you're just siblings born at the same time. The concept of having someone who shares my exact DNA code (of course, save for mutations that develop over a lifetime or differences in our epigenomes) is just really fascinating to me. Thus, it has always been important for me to know whether Matt and I are identical or fraternal twins.
Last May, my close friend Pat Nosker informed me about 23andMe, a personal genome sequencing service. 23andMe was founded in April 2006 and has attracted significant investment from multiple companies, including Google (one of 23andMe's co-founders, Anne Wojcicki, is married to Google co-founder Sergey Brin). Pat and I purchased kits from 23andMe and had a lot of fun going over our results, and several months later I convinced my twin brother Matt to purchase his own kit. Also, earlier this year I contacted 23andMe about receiving a free upgrade kit for the purposes of this article and they graciously provided me one, so many thanks to them for letting me get the most updated data. With my results already in hand, once Matt received his we would finally find out if we are identical or fraternal twins.
Before Matt and I could determine whether we are identical or fraternal twins, we had to complete our 23andMe kits. The main goal is to get some of our DNA into a preservation vial before shipping it back to the 23andMe sequencing lab. In order to get a sample of DNA, the kit instructions tell each user to swish around saliva in his or her mouth before gently spitting it into the perservation vial. In doing this, we are removing some of our inner cheek cells, similar to a buccal swab. After about five minutes of swishing and spitting, there is enough saliva (and therefore cells) in the preservation vial. In order to preserve the sample, users then close the cap, which punctures a container of preservation fluid and deposits it in to the saliva sample. After a few quick shakes, the sample is placed in a biohazard bag and ready to be shipped back to the 23andMe sequencing lab in California.
Contents of the 23andMe kit.

Preservation vial before use. The preservation fluid is contained within the large blue cap.
Preservation vial with complete saliva sample and preservation fluid all mixed up.
23andMe and similar personal genome service companies including Navigenics and deCODEme don't sequence your entire genome. In fact, they don't even come close: out of the 3 billion base pairs present in the human genome, 23andMe only sequences around 950,000, or just about 0.032%. What 23andMe sequences are single nucleotide polymorphisms, or SNPs. SNPs are defined as DNA sequence variations at the individual base pair level. Variance of single base pairs across our genome are what make us unique, and they are also the causes of many genetic diseases, including the disease I study in graduate school, fibrodysplasia ossificans progressiva (FOP). Different bases at a particular spot in our DNA sequence can cause an amino acid change in our proteins, and if these mutations occur within an important and evolutionarily conserved active region of the protein, as is the case with FOP, then there can be drastic effects for the organism.
Even though personal genome services sequence a very small portion of your genome, you are still able to learn much about your individual DNA code. The overwhelming majority of our genome (~96-97%) does not code for proteins, and while this non-coding DNA is far from junk, mutations in this portion of the DNA are generally more tolerated than in our evolutionarily conserved regions. Therefore, sequencing companies have focused on sequencing SNPs in our coding DNA regions first. However, the costs for sequencing our entire genome are decreasing every year, and currently it is pretty reasonable, at least in the scope of funded research labs (around $10,000, and perhaps even less). Experts are predicting that within another 5-10 years, it will be cost effective (around ~$100-200 per person) to sequence everyone's entire genome, and companies like 23andMe, Navigenics, and deCODEme will have to evolve their lab facilities and business models to adjust to changes in the field.
Now that I have explained how to complete a 23andMe kit, as well as how it is processed in the lab, I'm going to let you know how my results turned out.
After a few weeks of waiting, I finally received my results. Turns out I'm pretty normal. I have an increased risk for prostate and colon cancer, though not excessively so. In contrast, I have a decreased risk for melanoma and rheumatoid arthritis. Other than those diseases and a couple more, I'm actually within the average for the most of the reported diseases, which is encouraging. However, most of these diseases can be caused by multiple genetic defects, so there always is a bit of uncertainty when it comes to definitively knowing whether an individual will develop symptoms. For other diseases there is much more absolute genetic linkage (termed penetrance by geneticists). These diseases are listed in the "Carrier Status" section, and some of the more serious ones (like Cystic Fibrosis) are actually locked even after electronically signing the "you might learn things you don't want to know about yourself" waiver! I chose to unlock all of the diseases in this section, and luckily I don't have any of the most common SNPs associated with the serious diseases (though, it's important to note, that while lacking any of the common SNPs associated with a disease very likely means you don't have the disease, it isn't 100% certainty, as you may have mutations in the gene that are not sequenced). Interestingly, I was positive as a carrier for a SNP associated with mild hemochromatosis, or excessive iron levels. Thankfully the disease is recessive, so I need both copies to present symptoms, but this information is useful for future family planning.
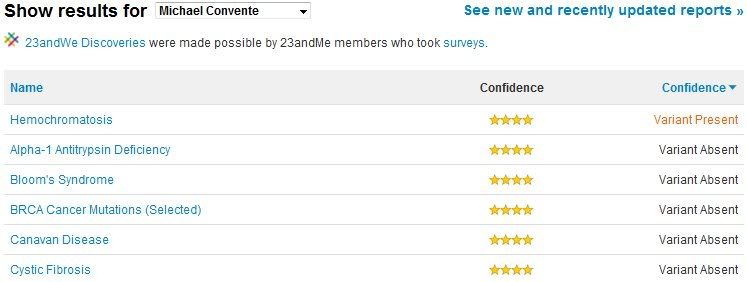
Listed above are some of the diseases assesed in the SNP sequencing done by 23andMe. Based on my report, I learned that I am a carrier for a mutation causing hemochromatosis.
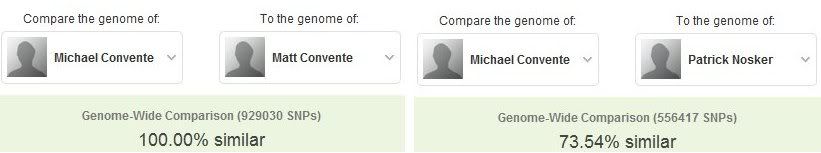
Though I learned a lot about my disease risks and carrier statuses in my report, I was most interested in finally learning whether I am an identical or fraternal twin. After my brother received his results in January 2011, I was elated to find out that we indeed are identical twins. 23andMe allows users to share their results with other members, and members who are linked are able to compare their SNPs, disease risk percentanges, and carrier statuses. As you can see below, Matt and I are 100% similar in our SNPs, whereas Pat and I are only 73.54% similar, which is actually a few percentage points higher than the average human.
One of the cool things about 23andMe is the ability for users to download their raw data. Since the large majority of SNPs sequenced by 23andMe are not associated with disease (or any known gene function for that matter) at high confidence, the information provided to users on their website covers a small portion of their entire SNP sequence. This is understandable as customers who may not be well versed in biology or genetics could become unnecessarily alarmed at possessing a SNP reported to be associated with a disease with very low penetrance. However, for customers who are more knowledgable in genetics and scientific research (like myself), having the raw data allows for additional reporting provided by third-party software. One of these third-party platforms is called Promethease, run by a wiki-formatted group called SNPedia. For a very reasonable $2 fee, you can upload your raw data file into the Promethease program and have it scan your SNPs for additional diseases or traits. Many of the disease reports are of lower confidence than reported by 23andMe, but it still is cool to have the most information possible.
The best feature of Promethease is the ability to run a visual comparison between your data and another person's data. The result is a visualization of your actual microarray data for each SNP sequenced. Comparing my chromosome 1 SNPs to my twin brother Matt's, you can see that we are almost the same for the entire array matrix (light green means identical SNPs for both copies of the chromosome; i.e. - the copy from mom and the copy from dad). There are some dark green spots, which represent a difference on only one chromosome, but there are so few that it they are likely due to sequencing or readout errors. In contrast, there are many more differences when comparing my chromosome 1 SNPs to Pat's (red spots represent differences on both copies of the chromosome).
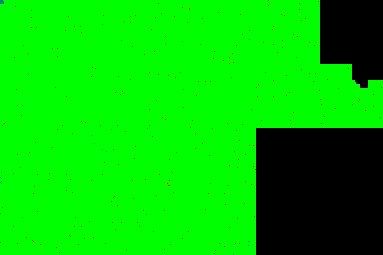

Top: Comparison of Chromosome 1 between Matt and Mike. Bottom: Comparison of Chromosome 1 between Pat and Mike.
Overall, as a result of having my DNA sequenced via a personal genome service I have learned for certain that I am an identical twin as well as some interesting information about being a carrier for a mutation causing mild hemochromatosis. For me, confirming that I am indeed an identical twin was well worth the cost of the service, and any additional knowledge I gained from it was a bonus. I definitely learned a lot of information, but there are definite limitations to the service, and it may not be for everyone. Lastly, I will offer some commentary about the future of personal genomics and how it can still be improved.
It's clear that we obtain a lot of data after sequencing just under 1 million SNPs. However, the biggest impediment to the field of personal genomics is knowing what to do with all the data we obtain. There are a solid number of diseases from which we can say with certainty whether someone will develop them. However, for the vast majority of diseases, there not only are multiple genes that can be involved, but multiple SNPs in a single gene. This makes analysis of an individual's genome potentially quite difficult. Add to the mix a relative information gap of genetics and biology knowledge among the general population, and the final result is often confusion. The ability to sequence our entire genome at low cost will only make the problem worse, as sequencing technology is getting more advanced and cheaper far quicker than researchers are uncovering links between certain SNPs and disease.
Lastly, there are serious bioethical implications about knowing your genome sequence. Would you want to know for certain that you would get cystic fibrosis or Alzheimer's disease at some point in the future? Some of the most deleterious diseases are unfortunately also are genetically diagnosed with utmost certainty. Knowing with certainty at a young age a serious disease is in your future could strongly impact life decisions, such as starting a family. If I had a family history of a certain disease, personally I would want to know. There is no greater truth than our DNA, so in my opinion it's better to be prepared than to be uninformed. These important questions will no doubt start to enter into our everyday family and medical discussions. The field of personal genomics is still in it's early stages, but it is definitely here to stay.


